ML for protein engineering seminar series
A bi-weekly seminar series focused on recent work in machine learning for protein engineering.
For information on this seminar series and upcoming speakers, visit our website and twitter!
https://www.ml4proteinengineering.com/
https://twitter.com/ml4proteins
All seminars will be recorded and released on YouTube following the live Zoom session.

Early Career Seminar Series #2: Prof. Noelia Ferruz

Early Career Seminar Series #1: Prof. Mohammed AlQuraishi

Deep learning guided design of protease substrates

Atom level enzyme active site scaffolding using RFdiffusion2
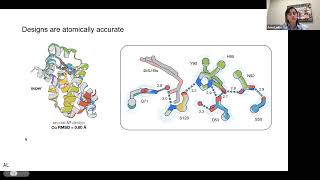
Computational design of serine hydrolases

Learning millisecond protein dynamics from what is missing in NMR spectra

MotifBench: A standardized protein design benchmark for motif-scaffolding problems

Genome modeling and design across all domains of life with Evo 2

Targeting protein–ligand neosurfaces with a generalizable deep learning tool

Bayesian Optimization of Antibodies Informed by a Generative Model of Evolving Sequences

ProtComposer: Compositional Protein Structure Generation with 3D Ellipsoids

Proteina: Scaling Flow-based Protein Structure Generative Models

Concept Bottleneck Language Models for Protein Design

InterPLM: Discovering Interpretable Features in Protein Language Models via Sparse Autoencoders

Mapping the combinatorial coding between olfactory receptors and perception with deep learning

Multistate and functional protein design using RoseTTAFold sequence space diffusion

ProTrek: Navigating the Protein Universe through Tri-Modal Contrastive Learning

AI-driven de novo Enzyme Design

The OMG dataset: An Open MetaGenomic corpus for mixed-modality genomic language modeling

Template-based protein editing using Raygun

Lessons from implementing AlphaFold3 in the wild

Tokenized and Continuous Embedding Compressions of Protein Sequence and Structure (CHEAP)

Adapting protein language models for structure-conditioned design

Rapid protein evolution by few-shot learning with a protein language model

Sequence-Augmented SE (3)-Flow Matching For Conditional Protein Backbone Generation

CARE: a Benchmark Suite for the Classification and Retrieval of Enzymes

Simulating 500 million years of evolution with a language model

Engineering highly active and diverse nuclease enzymes by ML and high-throughput screening

Designing and Scaffolding Proteins at the Scale of the Structural Universe with Genie 2

Framework for conditional diffusion models with applications in motif scaffolding for protein design